A biosynthetic pathway was designed that began with bacterial synthesis of the backbone structure, N-acetyl heparosan, with the aim of producing a bioengineered heparin product. N-Acetyl heparosan is synthesized by E. coli K5 as a capsular polysaccharide and is shed into the growth media.
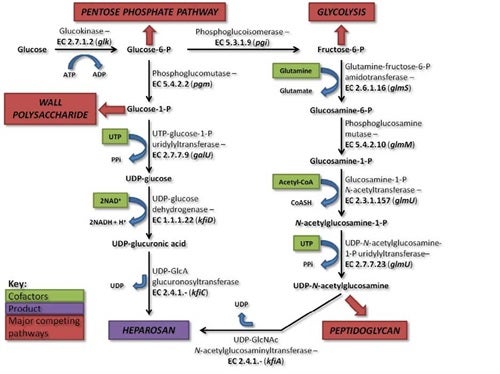
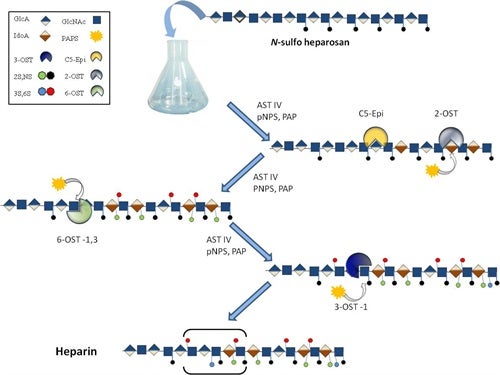
Once extracted and purified, the heparosan polysaccharide undergoes a series of in vitro chemoenzymatic modifications to make heparin, with the initial steps (N-deactylation and N-sulfonation) involving chemical modification and the final steps (epimerization of glucuronic acid to iduronic acid, 2-O-, 6-O-, and 3-O-sulfation, involving enzymatic modification. Since heparin production would be reliant on microorganisms, instead of animals, scale up could be achieved by increasing fermentation volume of the heparosan producer, E. coli K5, and the recombinant E. coli strains used to produce the heparosan modifying enzymes. The process has been completed on a small scale and is being optimized in order to pursue heparin synthesis on a larger scale that could eventually be scaled up to produce bioengineered heparin at an industrial scale.
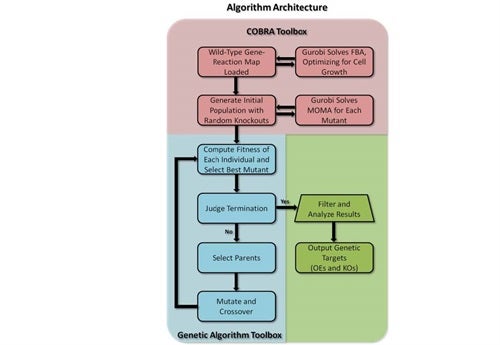
In a related project, in collaboration with the Koffas lab, we have recently sequenced the genome of Escherichia coli K5 and have constructed a draft genome-scale metabolic reconstruction to improve the endogenous K5 capsular polysaccharide (heparosan) production capacity of this E. coli strain using stoichiometric-based modeling. A multilevel optimization algorithm capable of predicting combinations of genetic manipulations for increased production of target metabolites has been constructed by building upon the success of Cipher of Evolutionary Design (CiED), a genetic algorithm developed in the Koffas lab for predicting gene knockouts.
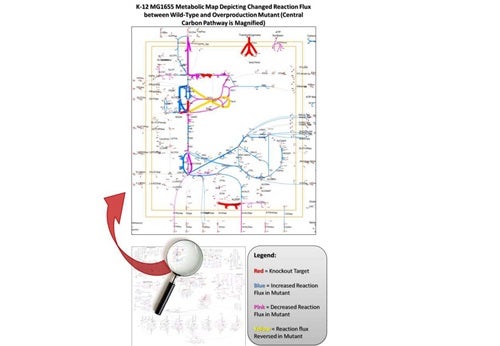
Application of the MATLAB-based algorithm to the newly constructed metabolic model allows prediction of sets of gene overexpressions and deletions consistent with the desired cellular phenotype, high heparosan production levels. Computational predictions for improving availability of heparosan precursors are consistent with previous experimental results and are being evaluated in the lab.
We are also utilizing traditional metabolic engineering approaches aimed at increasing intracellular supply of heparosan precursors, such as overexpression of the capsular polysaccharide gene cluster, overexpression of enzymes in the metabolic pathway, and elimination of nonessential competing pathways by targeted gene deletion.
Mutant E. coli K5 strains possessing a combination of rational and predicted genetic manipulations have the potential to achieve significantly improved heparosan titers, making E. coli a safe and viable heparosan production platform as a source for bioengineered heparin.
